Current Research
1. The Enzymatic Mechanism of Oxalate Decarboxylase.
1. The Enzymatic Mechanism of Oxalate Decarboxylase. Oxalic acid is the simplest dicarboxylic acid but also the most common in our environment. It is produced by plants and is the most common naturally occuring toxin in our diet. It is particularly common in big-leaf edible plants such as rhubarb, chard, beet, and spinach. Our kidneys have to flush out excess oxalate and if we ingest too much we end up with kidney stones or even worse consequences. Oxalate decarboxylase (OxDC) is one of only three known enzymes that degrade oxalic acid. It is common in soil bacteria and fungi and plays an important role in the biogeochemical carbon cycle. OxDC breaks down oxalate by a redox-neutral uni-molecular disproportionation reaction according to the following chemical reaction: HC2O4 (aq) → CO2 + HCO2 (aq) The reaction mechanism is still poorly understood. We utilize electron paramagnetic resonance (EPR) to study the manganese ions contained in the enzyme and follow their redox states under reaction turnover conditions. We also test for free radicals produced in this reaction. Enzymatic assays, X-ray crystallography, mass spectrometry combined with site-directed mutagenesis of the protein are further approaches we take to this problem. Our research has contributed the following important results to the knowledge about OxDC: (1) During turnover the protein generates the superoxide free radical, O2−∙. (2) The presence of Mn(III) was observed by EPR spectroscopy and its pH dependence matches that of the enzyme activity suggesting that Mn(III) drives the catalytic process (see figure). (3) We were able to obtain the first crystal structure of OxDC at low pH in the active state. This has led us to propose a new model for substrate binding.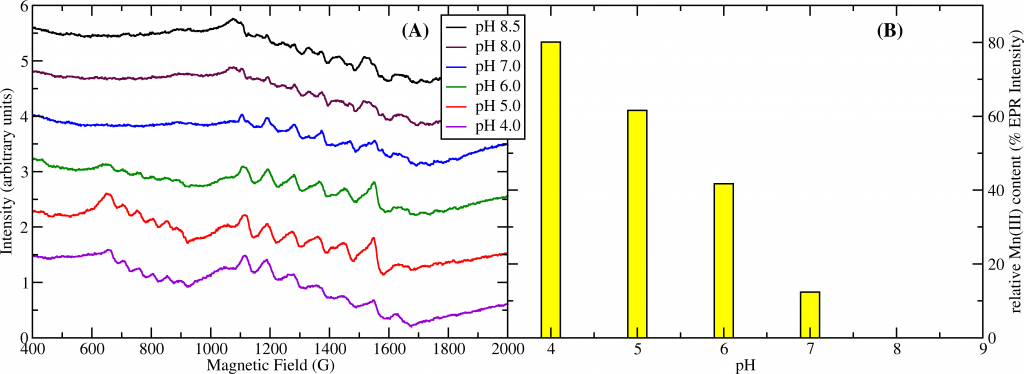
Figure 1: (A) pH titration of OxDC. The sextet signal between 600 and 900 G corresponds to Mn(III) while the signals between 1100 and 1700 G correspond to Mn(II). (B) relative concentration of Mn(III) based on its EPR intensity. Mn(III) has maximum intensity at pH4 while there is none detectable at pH8. This correlates strongly with the pH dependence of the enzyme activity which also peaks at pH4 and is absent at pH8.